Genetic diversity of Phytophthora nicotianae reveals pathogen transmission mode in Japan
- 79 Downloads
Abstract
Phytophthora nicotianae is an important soil-borne pathogen in tropical, subtropical and temperate regions. To clarify the genetic diversity of P. nicotianae and to understand its mode of transmission in Japan, we developed six new microsatellites markers, consisting of six loci and 39 alleles. In a phylogenetic analysis, 138 isolates, including 125 from Japan and 13 from overseas, were shown to differ, even though some were collected from the same host and location, suggesting that there is no geographic or host plant clustering. Population structure analysis also revealed a highly admixed population of P. nicotianae in Japan. Molecular analysis suggested high variance between individuals but no significant differences between populations. Both A1 and A2 mating types were present in the same population, which could be due to high levels of variance between individuals in the population. The absence of geographical structure between populations also suggests that the pathogen is able to migrate from one population to another. We propose that this phenomenon could result from human activities related to the transport of plant and associated agricultural materials.
Keywords
Diversity Microsatellite Phytophthora nicotianae Population genetics Population structureIntroduction
As advancements in transportation technology have made global trading easier, the resultant global redistribution of species by human activities has included not only the introduction of beneficial species to new environments, but also the introduction of their associated pathogens. Most crops are in the process of rapid biotic homogenization, which can potentially lead to significant reductions in the genetic variability of the principal crops of many important agricultural nations within the next few decades (Bebber et al. 2014). Such losses pose a threat to crop species because the evolution of a new virulent variant of a pathogen could result in high losses and poor yields if the pathogen spreads rapidly. The near extinction of the Gros Michel banana in the 1950s is a good example: the lack of genetic variation within the banana population made it highly susceptible to a new strain of Fusarium that causes Panama disease. Genetic variability within a population has a direct impact on the virulence and ecology of certain pathogens because a highly variable gene pool allows them to adapt quicker to environmental change, thus increasing their potential to produce new virulent variants.
The oomycete, P. nicotianae, first isolated by De Haan in 1896, is one of the most devastating oomycete plant pathogens in the world because its broad host range includes over 255 species across a wide diversity of climates around the world (Panabières et al. 2016). P. nicotianae was first reported in Japan in 1934 when it was isolated from Agapanthus seedlings with leaf blight by Takimoto and blight of lily by Tasugi and Kumazama (Asuyama 1934). At that time, P. nicotianae was reported under the name P. parasitica, which is now considered to be a synonym (Cline et al. 2008). Major outbreaks of P. nicotianae in Japan have caused root rot of strawberries (Matsuzaki 1988; Suzui et al. 1980). More recent reports of P. nicotianae in Japan have included a broad range of host plants such as poinsettia (Kanto et al. 2007), passion fruit (Horie 2007), citrus (Tashiro et al. 2002), asparagus (Yokota et al. 2013), Welsh onion (Takeuchi and Suzuki 2010), kalanchoe (Watanabe et al. 2007), New Zealand spinach (Takeuchi et al. 2004), garden pea (Takeuchi and Horie 2000) and Limonium (Nakamura and Matsuzaki 1994).
Population genetic studies of P. nicotianae have mainly focused on isolates from tobacco (Bonnet et al. 1994; Colas et al. 1998; Mammella et al. 2013). Recent analysis, using single nucleotide polymorphisms (SNPs) on mitochondrial and nuclear genes, grouped the isolates based on their host plants (Mammella et al. 2013). However, isolates from nurseries exhibit less association between the host plant and genetic grouping (Biasi et al. 2016). The absence of geographic structure for P. nicotianae revealed a recent expansion of a single diverse population (Bruberg et al. 2011).
Various types of genetic markers, including mitochondrial DNA (mtDNA), random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), single nucleotide polymorphism (SNP) and microsatellites, are widely used in the study of population genetics. Factors such as the level of variability or marker sensitivity, the nature of the marker (e.g., dominant or co-dominant, multilocus or single locus) and available equipment need to be considered when selecting the most suitable marker for population genetic analysis (Sunnucks 2000). On the basis of these factors, we opted to use microsatellites for this population genetics study.
Microsatellites, often also referred to as simple sequence repeats (SSRs), are tandemly repeating units of DNA with a repeat size of 1–6 bp, flanked by regions of non-repetitive unique DNA sequences. Microsatellites are very sensitive markers with a high level of variability within their repeat sequence, which means that they can be used to detect alleles at a locus. They usually have a high mutation rate because they are in a noncoding region. Moreover, inheritance of microsatellites alleles is Mendelian. All these advantages make microsatellites an excellent genetic marker for high-resolution population analysis (Selkoe and Toonen 2006).
Data on population structure can help us gain a better understanding of the genotypic diversity among and within a population. The genetic structure of the pathogen population can affect the genetic resistance of that pathogen. The more genetically diverse a population, the more likely that the population will survive in threatening environments (Charlesworth 2015). In this study, we needed to develop a reliable microsatellite marker to obtain a robust and comprehensive data set on population structure. Due to the importance of understanding population genetics for disease management strategies, the objectives of this study were to (1) develop microsatellite markers that are reliable for P. nicotianae population genetic analysis and (2) summarize the genetic diversity of P. nicotianae in Japan.
Materials and methods
Phytophthora nicotianae isolates
We evaluated 138 isolates of P. nicotianae: 125 isolates from 38 host plants across 15 prefectures in Japan, four isolates from Taiwan, two from the United States, and seven from Indonesia (Table 1). Some isolates were obtained from the culture collections of Gifu University, Japan Ministry of Agriculture, Forestry, and Fisheries (MAFF) and National Institute of Technology and Evaluation Japan (NITE) Biological Research Center (NBRC) and others were isolated for this study from infected pineapple and tobacco plants in Indonesia (Table 1). To investigate local population dynamics, we collected 23 isolates from kalanchoe fields in Gifu (2004–2009) and 16 from strawberry and asparagus fields in Saga (2012–2013).
Isolates of Phytophthora nicotianae used in this study
| Working number | Population | Isolates | Host plant | Geographical origin | Isolation year | Mating type |
|---|---|---|---|---|---|---|
| 1 | Chubu | MAFF 712194 | Periwinkle (Catharanthus roseus) | Aichi, Japan | 1997 | A2 |
| 2 | Chubu | GK10NI2SH | Periwinkle (Catharanthus roseus) | Gifu, Gifu, Japan | 2010 | A1 |
| 3 | Chubu | GK 08NI8S2 | Periwinkle (Cattaranthus roseus) | Gifu, Gifu, Japan | 2008 | A1 |
| 4 | Chubu | GK10NI1SH | Periwinkle (Catharanthus roseus) | Gifu, Gifu, Japan | 2010 | A1 |
| 5 | Chubu | OINB113SH | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2004 | A2 |
| 6 | Chubu | OINB153 | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2004 | A2 |
| 7 | Chubu | OINB172 | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2004 | A2 |
| 8 | Chubu | OINB171SH | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2004 | A2 |
| 9 | Chubu | OINB171 | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2004 | A2 |
| 10 | Chubu | OINB153SH | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2004 | A2 |
| 11 | Chubu | OINB 113 | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2004 | A2 |
| 12 | Chubu | OINB 161 | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2004 | A2 |
| 13 | Chubu | OIOL0591R | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2005 | A2 |
| 14 | Chubu | OINO 451 SH | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2005 | A2 |
| 15 | Chubu | OINO 451 | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2005 | A2 |
| 16 | Chubu | OIOLO 0581 R | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2005 | A2 |
| 17 | Chubu | 0705W21 | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2007 | A2 |
| 18 | Chubu | 0805WJ21 | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2008 | A1 |
| 19 | Chubu | 09E11294 | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2009 | A1 |
| 20 | Chubu | 09E321 SH | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2009 | A2 |
| 21 | Chubu | 09W422 SH | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2009 | A2 |
| 22 | Chubu | 09WCRS1-1 | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2009 | A2 |
| 23 | Chubu | 09W221 SH | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | 2009 | A2 |
| 24 | Chubu | PHK6214 SH | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | Unknown | A2 |
| 25 | Chubu | GK 4-11 | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | Unknown | A2 |
| 26 | Chubu | PHKq-11 | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | Unknown | A2 |
| 27 | Chubu | PGS1 SH | Kalanchoe (Kalanchoe sp.) | Katagata, Gifu, Japan | Unknown | A2 |
| 28 | Chubu | MAFF 712342 | China doll (Radermachera sinica) | Ise, Mie, Japan | Unknown | A2 |
| 29 | Chubu | NBRC 30595 | Strawberry (Fragaria xananassa) | Shizuoka, Japan | 1979 | nr |
| 30 | Chubu | MAFF 305926 | Strawberry (Fragaria xananassa) | Shizuoka, Japan | Unknown | A2 |
| 31 | Chubu | GF468 | Strawberry (Fragaria xananassa) | Gifu, Japan | 2003 | A2 |
| 32 | Chubu | GF524 | Rose of Sharon (Hibiscus syriacus) | Ogaki, Gifu, Japan | 2003 | A2 |
| 33 | Kansai | CH00POIN 2 | Poinsettia (Euphorbia pulcherrima) | Hyogo, Japan | 2000 | A2 |
| 34 | Kansai | CH00POIN3 | Poinsettia (Euphorbia pulcherrima) | Hyogo, Japan | 2000 | A2 |
| 35 | Kansai | MAFF 239554 | Poinsettia (Euphorbia pulcherrima) | Hyogo, Japan | 2003 | A2 |
| 36 | Kanto | CH08DAV11 | Euphorbia sp. | Chiba, Japan | 2008 | A2 |
| 37 | Kanto | C23 | Indian mallow (Abutilon sp.) | Chiba, Japan | 2007 | A2 |
| 38 | Kanto | C24 | Indian mallow (Abutilon sp.) | Tateyama, Chiba, Japan | 2007 | A2 |
| 39 | Kanto | MAFF305795 | African violet (Saintpaulia goetzeana) | Tachikawa, Tokyo, Japan | 1987 | A2 |
| 40 | Kanto | CH94AROE1 | Aloe vera | Miyoshi, Chiba, Japan | 1994 | A2a |
| 41 | Kanto | CH94AROE3 | Aloe vera | Miyoshi, Chiba, Japan | 1994 | A2a |
| 42 | Kanto | CH92ALS11 | Peruvian lily (Alstroemeria sp.) | Kyonan, Chiba, Japan | 1992 | A2a |
| 43 | Kanto | CH92ALS21 | Peruvian lily (Alstroemeria sp.) | Kyonan, Chiba, Japan | 1992 | A2a |
| 44 | Kanto | GUGC5631 | Peruvian lily (Alstroemeria sp.) | Kyonan, Chiba, Japan | 1992 | A2a |
| 45 | Kanto | CH93ANE1 | Spanish marigold (Anemone coronaria) | Kimitsu, Chiba, Japan | 1993 | A1a |
| 46 | Kanto | CH93ANE2 | Spanish marigold (Anemone coronaria) | Kimitsu, Chiba, Japan | 1993 | A1a |
| 47 | Kanto | CH 90-4 | Zebra plant (Aphelandra squarrosa) | Chiba, Chiba, Japan | 1990 | A2a |
| 48 | Kanto | CH90-9 | Zebra plant (Aphelandra squarrosa) | Chiba, Japan | 1990 | A2a |
| 49 | Kanto | CH90-6 | Zebra plant (Aphelandra squarrosa) | Chiba, Japan | 1990 | A2a |
| 50 | Kanto | CH89-44 | Bougenvillea sp. | Kyonan, Chiba, Japan | 1989 | A2a |
| 51 | Kanto | CH89-43 | Bougenvillia sp. | Kyonan, Chiba, Japan | 1989 | A2a |
| 52 | Kanto | C38 | Brodiaea sp. | Chiba, Japan | 2007 | A2 |
| 53 | Kanto | MAFF 305796 | Periwinkle (Cathtaranthus roseus) | Tokyo, Japan | 1988 | |
| 54 | Kanto | CH98Y1A | Yuzu (Citrus junos) | Futtsu, Chiba, Japan | 1998 | A1a |
| 55 | Kanto | CH98U1A | Tangerine (Citrus unshiu) | Futtsu, Chiba, Japan | 1998 | A1a |
| 56 | Kanto | MAFF 235436 | Daphne sp. | Ibaraki, Tsukuba, Japan | 1983 | Nr |
| 57 | Kanto | CH95PHJ2 | Winter daphne (Daphne odora) | Asahi, Chiba, Japan | 1995 | A2a |
| 58 | Kanto | CH95PHJ1 | Winter daphne (Daphne odora) | Asahi, Chiba, Japan | 1995 | A2a |
| 59 | Kanto | CH87CWE1 | Dianthus sp. | Wada, Chiba, Japan | 1987 | A2a |
| 60 | Kanto | CH87-51 | Dianthus sp. | Chikura, Chiba, Japan | 1987 | A2a |
| 61 | Kanto | GUGC5562 | Dianthus sp. | Chikura, Chiba, Japan | 1987 | A2a |
| 62 | Kanto | CH87KTK1 | Carnation (Dianthus caryophyllus) | Tomiura, Chiba, Japan | 1987 | A2a |
| 63 | Kanto | CH87WG1 | Carnation (Dianthus caryophyllus) | Wada, Chiba, Japan | 1987 | A2a |
| 64 | Kanto | CH87CWG1 | Carnation (Dianthus caryophyllus) | Wada, Chiba, Japan | 1987 | A2a |
| 65 | Kanto | CH87-50 | Dianthus sp. | Chiba, Japan | 1987 | A2a |
| 66 | Kanto | C15 | Echium fastuosum | Tateyama, Chiba, Japan | 2006 | A2 |
| 67 | Kanto | C58 | Gerbera sp. | Chiba, Japan | 2008 | A2 |
| 68 | Kanto | CH96HE1 | English ivy (Hedera helix) | Kyonan, Chiba, Japan | 1996 | A2a |
| 69 | Kanto | CH97HE11 | English ivy (Hedera helix) | Maruyama, Chiba, Japan | 1997 | A2a |
| 70 | Kanto | CH96HE2 | English ivy (Hedera helix) | Kyonan, Chiba, Japan | 1996 | A2a |
| 71 | Kanto | C26 | Lavender (Lavandula angustifolia) | Chiba, Japan | 2007 | A2 |
| 72 | Kanto | CH99LK1 | Lily (Lilium hybrida) | Kyonan, Chiba, Japan | 1999 | A2a |
| 73 | Kanto | CH91KK4 | Easter lily (Lilium longiflorum) | Kyonan, Chiba, Japan | 1991 | A2a |
| 74 | Kanto | GUGC5567 | Easter lily (Lilium longiflorum) | Kyonan, Chiba, Japan | 1991 | A2a |
| 75 | Kanto | GUGC5630 | Limonium sp. | Maruyama, Chiba, Japan | 1991 | A2a |
| 76 | Kanto | GUGC5673 | Limonium sp. | Maruyama, Chiba, Japan | 1991 | A2a |
| 77 | Kanto | CH91-33 | Limonium sp. | Maruyama, Chiba, Japan | 1991 | A2a |
| 78 | Kanto | CH91-29 | Limonium sp. | Maruyama, Chiba, Japan | 1991 | A2a |
| 79 | Kanto | CH92ORN21 | Ornithogallum sp. | Futtsu, Chiba, Japan | 1992 | A2a |
| 80 | Kanto | CH92ORN11 | Ornithogallum sp. | Futtsu, Chiba, Japan | 1992 | A2a |
| 81 | Kanto | CH93ORN4 | Ornithogallum sp. | Tateyama, Chiba, Japan | 1993 | A2a |
| 82 | Kanto | GUGC5632 | Ornithogallum sp. | Futtsu, Chiba, Japan | 1992 | A2a |
| 83 | Kanto | MAFF 712287 | Viola tricolor | Saitama, Japan | 2006 | A1 |
| 84 | Kanto | CH85PHP37 | Petroselinum crispum | Maruyama, Chiba, Japan | 1985 | A2a |
| 85 | Kanto | CH85PHP61 | Petroselinum crispum | Maruyama, Chiba, Japan | 1985 | A2a |
| 86 | Kanto | CH075STR81 | Strawberry (Fragaria xananassa) | Chiba, Japan | 2007 | A2a |
| 87 | Kanto | CH91-1 | Strelitzia sp. | Tateyama, Chiba, Japan | 1991 | A2a |
| 88 | Kanto | CH91-4 | Strelitzia sp. | Tateyama, Chiba, Japan | 1991 | A2a |
| 89 | Kanto | CH91-3 | Strelitzia sp. | Tateyama, Chiba, Japan | 1991 | A2a |
| 90 | Kanto | CH91-2 | Strelitzia sp. | Tateyama, Chiba, Japan | 1991 | A2* |
| 91 | Kanto | GUGC5633 | Strelitzia sp. | Chiba, Chiba, Japan | 1991 | A2* |
| 92 | Kanto | MAFF 305939 | Nicotiana rustica | Kanagawa, Japan | Unknown | Nr |
| 93 | Kanto | CH89-39 | Vanda sp. | Tateyama, Chiba, Japan | 1989 | A2a |
| 94 | Kanto | CH89-40 | Vanda sp. | Tateyama, Chiba, Japan | 1989 | A2a |
| 95 | Kanto | CH99TK2 | Lily (Lilium hybrida) | Chiba, Japan | 1999 | Nr |
| 96 | Kyushu | SG12ASP1-1 | Asparagus (Asparagus officinalis) | Saga, Japan | 2012 | A2 |
| 97 | Kyushu | SG12ASP1-2 | Asparagus (Asparagus officinalis) | Saga, Japan | 2012 | A2 |
| 98 | Kyushu | SG12ASP2-1 | Asparagus (Asparagus officinalis) | Saga, Japan | 2012 | A2 |
| 99 | Kyushu | SG12ASP1-3 | Asparagus (Asparagus officinalis) | Saga, Japan | 2012 | Nr |
| 100 | Kyushu | SG12ASP2-2 | Asparagus (Asparagus officinalis) | Saga, Japan | 2012 | A2 |
| 101 | Kyushu | SG13ASP1-2 | Asparagus (Asparagus officinalis) | Saga, Japan | 2013 | A2 |
| 102 | Kyushu | SG13ASP1-1 | Asparagus (Asparagus officinalis) | Saga, Japan | 2013 | A2 |
| 103 | Kyushu | SG13ASP1-3 | Asparagus (Asparagus officinalis) | Saga, Japan | 2013 | A1 |
| 104 | Kyushu | MAFF 237653 | Strawberry (Fragaria xananassa) | Saga, Japan | 1978 | A2 |
| 105 | Kyushu | MAFF 242197 | Strawberry (Fragaria xananassa) | Saga, Japan | 2004 | A2 |
| 106 | Kyushu | SGPC 0503 | Strawberry (Fragaria xananassa) | Saga, Japan | Unknown | A2 |
| 107 | Kyushu | SGPY 2101 | Strawberry (Fragaria xananassa) | Saga, Japan | Unknown | A2 |
| 108 | Kyushu | SGPC 0502 | Strawberry (Fragaria xananassa) | Saga, Japan | Unknown | A2 |
| 109 | Kyushu | SGPC 04118 | Strawberry (Fragaria xananassa) | Saga, Japan | Unknown | A2 |
| 110 | Kyushu | SGPC 0501 | Strawberry (Fragaria xananassa) | Saga, Japan | Unknown | A2 |
| 111 | Kyushu | SGHP0002 | Strawberry (Fragaria xananassa) | Saga, Japan | Unknown | A2 |
| 112 | Kyushu | MAFF 305940 | Nicotiana rustica | Kagoshima, Japan | 1977 | A2 |
| 113 | Kyushu | SE759 | na | Saga, Japan | A2 | |
| 114 | Kyushu | F03 | na | Fukuoka, Japan | 2006 | A1 |
| 115 | Shikoku | MAFF 238154 | Onion (Allium cepa) | Kochi, Japan | 1999 | A1 |
| 116 | Shikoku | NBRC 33191 | Scallion (Allium fistulosum) | Kochi, Japan | 1999 | A2a |
| 117 | Shikoku | NBRC 33190 | Scallion (Allium fistulosum) | Kochi, Japan | 1999 | A2a |
| 118 | Shikoku | MAFF 238152 | Lilium sp. | Kochi, Japan | 1999 | A2 |
| 119 | Shikoku | NBRC 33193 | Lilium sp. | Kochi, Japan | 1999 | A2a |
| 120 | Shikoku | NBRC 33192 | Flame lily (Gloriosa superba) | Kochi, Japan | 1999 | A2 |
| 121 | Southern Island | MAFF 305,797 | Dracaena sp. | Hachijojima, Tokyo, Japan | 1986 | Nr |
| 122 | Southern Island | MAFF 305591 | Papaya (Carica papaya) | Ogasawara, Tokyo, Japan | 1986 | A2 |
| 123 | Southern Island | MAFF 305799 | Passion fruit (Passiflora edulis) | Hachijojima, Tokyo, Japan | 1983 | A2 |
| 124 | Southern Island | MAFF 305978 | Passion fruit (Passiflora edulis) | Ogasawara, Tokyo, Japan | 1988 | A2 |
| 125 | Southern Island | MAFF 305590 | Tomato (Solanum lycopersicum) | Ogasawara, Tokyo, Japan | 1986 | Nr |
| 126 | Taiwan | NBRC 31425 | Onion (Allium cepa) | Taiwan | 1984 | A1a |
| 127 | Taiwan | NBRC 31423 | Pineapple (Annanas comosus) | Taiwan | 1984 | A1a |
| 128 | Taiwan | NBRC 31419 | Papaya (Carica papaya) | Taiwan | 1984 | A2a |
| 129 | Taiwan | NBRC 31416 | Tomato (Solanum lycopersicum) | Taiwan | 1984 | A2a |
| 130 | Indonesia | TBC GTS | Tobacco (Nicotiana rustica) | Central Java, Indonesia | 2016 | A1 |
| 131 | Indonesia | AA 129D 2 | Pineapple (Annanas comosus) | Lampung, Indonesia | 2016 | A1 |
| 132 | Indonesia | AA 71A S1 | Pineapple (Annanas comosus) | Lampung, Indonesia | 2016 | A2 |
| 133 | Indonesia | AA 114K HS 2 | Pineapple (Annanas comosus) | Lampung, Indonesia | 2016 | A1 |
| 134 | Indonesia | AA 71A 2 | Pineapple (Annanas comosus) | Lampung, Indonesia | 2016 | A2 |
| 135 | Indonesia | AA 36G | Pineapple (Annanas comosus) | Lampung, Indonesia | 2016 | A1 |
| 136 | Indonesia | AA 71A 3 | Pineapple (Annanas comosus) | Lampung, Indonesia | 2016 | A1 |
| 137 | USA | CBS 535.92 | Soil | USA | A1a | |
| 138 | USA | CBS 534.92 | Soil | USA | A2a |
Phytophthora nicotianae was isolated from infected plant tissues on selective NARM agar as previously described (Morita and Tojo 2007). The resultant mycelia were then identified by sequencing the internal transcribed spacer (ITS) region and the cytochrome c oxidase 1 (COX1) gene (Robideau et al. 2011). The isolates were categorized into nine population groups based on their geographical origin: five populations from the largest main island Honshu (Chubu, Kansai, Kanto, Kyushu, Shikoku) and the southern islands of Japan and three populations from overseas (Taiwan, USA, and Indonesia).
Mating type determination
Isolate mating types were determined as previously described (Parkunan et al. 2010). Unknown mating types were paired with known A1 and A2 isolates (CH92ALS11 and CH93ANE1, respectively) on V8 agar, then incubated until a mating zone formed and antheridia and oogonia were observed.
Microsatellite marker development
The complete genome sequence of P. nicotianae was screened for the microsatellite motifs using Tandem Repeat Finder (Benson 1999). The alignment parameters for Tandem Repeat Finder were 2, 3 and 5, and only those repeats with a minimum score of 80 and a maximum period size of 6 were reported. The microsatellites were selected on the basis of a minimum of three repeats for trinucleotides and tetranucleotides. Primers flanking the identified loci were designed, and their specificity was confirmed using Primer BLAST (Ye et al. 2012). All primers were designed using the following criteria: Tm of 55–65°C (optimum at 58°C), product size of 150–250 bp (optimum at 200 bp), GC content 45–60% (optimum at 50%) and primer size of 18–25 bp (optimum at 20 bp).
All primers were analyzed for hairpin and dimer potential using NetPrimer (http://www.premierbiosoft.com/NetPrimer/AnalyzePrimer.jsp) to select the best primer pairs. These selected primer pairs were then analyzed against the whole genome sequence of P. nicotianae by in silico PCR using the Web-based program insilico.ehu.eus (San Millán et al. 2013). Amplified fragments were cloned using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA, USA) and then sequenced to characterize their microsatellite motifs. More than 12 E. coli recombinants were selected by colony PCR and purified using the ExoSAP-IT kit, following the manufacturer’s instructions (Affimetrix, Santa Clara, CA, USA). The purified PCR product was sequenced using the M13M4 primer for amplification by the BigDye Sequence Terminator Kit (Applied Biosystems, Foster City, CA, USA) on an ABI3500 automated sequencer (Applied Biosystems).
Microsatellite genotyping
The developed polymorphic loci were used to analyze all 138 isolates. The primers were labeled at the 5′ end separately with the fluorescent dye FAM (6-carboxy-fluorescein) or HEX (4,7,2′,4′,5′,7′-hexachloro-6-carboxyfluorescein) (Lees et al. 2006).
The total genomic DNA was extracted using PrepMan Ultra Reagent (Applied Biosystem) and amplified using all selected primers under the following conditions: 1 cycle of 94°C for 5 min; 35 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 30 s; and a final extension at 72°C for 7 min. Reactions were performed in a total volume of 25 µl containing 2 µl of 1 ng DNA, 2.5 µl of 10 × PCR Buffer (plus magnesium, Takara Bio, Otsu, Shiga, Japan), 2.5 µl of 4 mg/ml BSA, 2.5 µl of 10 mM primer (forward and reverse), 2 µl of 2.5 mM dNTP mix (Takara Bio), 0.1 µl rTaq polymerase (Takara Bio), and 10.9 µl ddH2O. PCR amplification products were separated in 2% agarose gels in 0.5 × Tris-acetate-EDTA buffer, stained with GelRed (Biotium, Fremont, CA, USA) and visualized under UV light.
After confirmation of the PCR product, fragments were analyzed on an ABI3100 or ABI3130 Genetic Analyzer (Applied Biosystem) using the LIZ 250 DNA ladder as a marker. The electropherogram was scored manually.
Population structure analysis
In a cluster analysis of the population structure, the probability of genotypes being distributed into K number of clusters was estimated using structure v. 2.3.4. (Falush et al. 2003, 2007; Hubisz et al. 2009; Pritchard et al. 2000) with an admixture model without prior population information and 200,000 Markov chain Monte Carlo (MCMC) iterations. Eight independent runs were performed for each K = 1–20. The optimal number of K was selected by STRUCTURE HARVESTER (Earl and Von Holdt 2012) and matched from an independent run by CLUMPP (Jakobsson and Rosenberg 2007). The result was then finally visualized using distruct (Rosenberg 2004). The distant matrix created by GenAlex 5.6.3. (Peakall and Smouse 2006, 2012) was used for phylogenetic analysis using a neighbor-joining algorithm in MEGA 6.0 (Tamura et al. 2013). The mating pattern within the population was statistically analyzed using an analysis of molecular variance (AMOVA) in GenAlex 5.6.3 (Peakall and Smouse 2006, 2012).
Results
Development of microsatellite markers
The entire genome sequence was screened using Tandem Repeat Finder, and 12 primer sets were selected that could specifically amplify 12 microsatellite loci of P. nicotianae. Those primer sets were then tested on three isolates (GUCC 5620, 5621 and 5623), and the loci that had multiple alleles were selected for study (Table 2). The selection of microsatellite markers established six novel polymorphic microsatellite loci. Six of 12 selected primer sets were suitable for population structure analysis because they were amplified in all isolates, diploid, and highly polymorphic.
Novel microsatellite markers of Phytophthora nicotianae developed in this study
| Locus | Repeat motif | Primer sequence | Annealing temperature (°C) | Fluorescent label | Alleles | |
|---|---|---|---|---|---|---|
| N | Size | |||||
| AA-TTA | TTA | F: CGTGAGGCAGATGCTGTCAA R: TGGGTTTCAGCCCTTCAACT | 60 | FAM | 4 | 263–287 |
| AA-AAC | AAC | F: GAGTTCTACATCCCGGTTCCA R: GCTTATAGTGGTGCAAGCGTC | 60 | FAM | 10 | 193–220 |
| AA-GCT | GCT | F: CTGGACATGCTCAGGGTGTT R: GACTGGATGGATCCGGCTTG | 60 | FAM | 5 | 177–189 |
| AA-CAG | CAG | F: ACGACCCATTCGCTGTTCAA R: TTTCCGTTGTTTGTGGGTGC | 60 | HEX | 4 | 234–246 |
| AA-TAA | TAA | F: TCTACGTCAGGGCGGTTTTT R: GAAATGTGTGGGTCAGTCGC | 60 | HEX | 4 | 170–179 |
| AA-GAA | GAA | F: GTGTCTTCACTGTCACCGGCAGTAGAA R: GTGTCTTCGGTTGGTCCAAACCTCTCC | 60 | HEX | 5 | 282–294 |
In total, 39 alleles were detected from six loci, ranging from four (TAA) to 11 (GTA) alleles per locus, with an average of 6.5 (Table 3) and maximum of 11 at locus GTA. This locus was also the most informative, as it had the highest Shannon’s Information Index (I = 1.838). Two of six alleles had significantly higher observed heterozygosity, while the rest were significantly lower. All of the loci significantly differed from Hardy–Weinberg equilibrium (HWE).
Microsatellite characteristics
| Locus | Na | Ne | I | H o | H e | p | F ST |
|---|---|---|---|---|---|---|---|
| AA-GAA | 5 | 2.401 | 1.068 | 0.779 | 0.584 | 0 | 0.072 |
| AA-GTA | 11 | 5.167 | 1.838 | 0.717 | 0.806 | 0 | 0.199 |
| AA-AAC | 7 | 3.798 | 1.484 | 0.649 | 0.737 | 0 | 0.18 |
| AA-CAG | 7 | 1.865 | 0.837 | 0.462 | 0.464 | 0 | 0.061 |
| AA-TTA | 5 | 3.691 | 1.357 | 0.752 | 0.729 | 0.002 | 0.119 |
| AA-TAA | 4 | 1.246 | 0.442 | 0.137 | 0.197 | 0 | 0.111 |
Mating type distribution
From 138 isolates, 21 isolates were identified as mating type A1, 95 as A2, and 22 isolates had no reaction to either the A1 or A2 mating type. Both A1 and A2 mating types were found on one kalanchoe farm in Gifu (Japan), one asparagus farm (Saga), one onion field (Kochi), and one pineapple field (Lampung, Indonesia). On the kalanchoe and asparagus farms, the A1 and A2 mating types were isolated in different years, while both mating types were isolated the same year in the Indonesian pineapple field (Table 1).
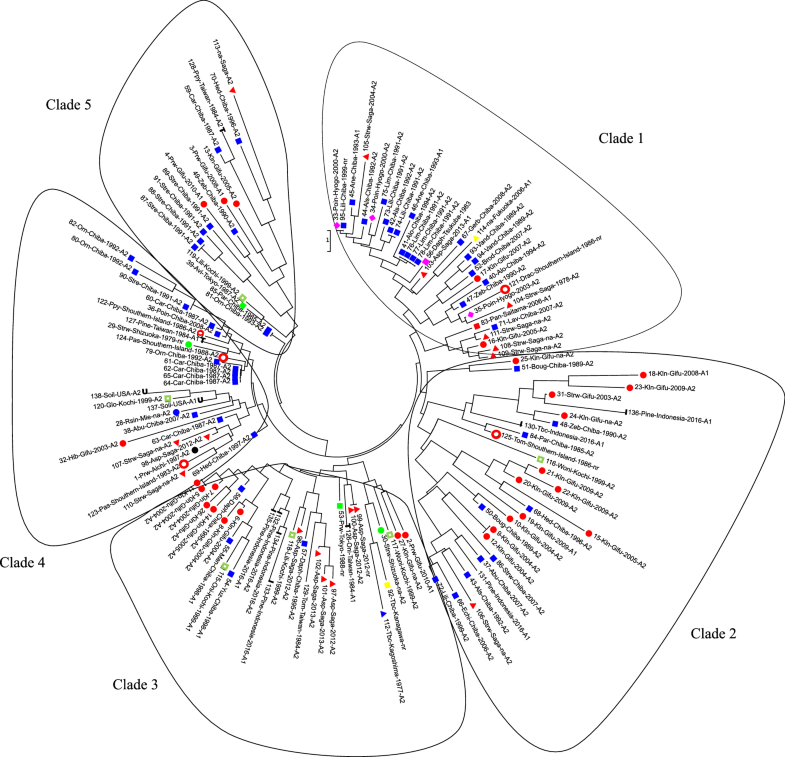
Phylogenetic analysis
The phylogenetic tree constructed with the neighbor joining algorithm revealed five major clades (Fig. 1). The isolates collected from the Kanto area were scattered in all clades in the phylogenetic tree as well as the isolates collected from Taiwan, Shikoku, and the southern islands. Isolates from the same geographic origin but different host species were found to be distantly related as were isolates from the same host species but different geographic origins. However, several isolates had the same genotype as other isolates collected from the same host and same geographic origin: three isolates (working number 76–78) from Limonium sp. in Chiba (clade 1), two isolates (No. 5 and 11) from Kalanchoe sp. in Gifu (Clade 2), two isolates (No. 9 and 12) also collected from Kalanchoe sp. in Gifu (Clade 3), two isolates (No. 9 and 100) from asparagus in Saga (Clade 3), four isolates (No. 61–64) from carnation in Chiba (Clade 4), and two isolates (No. 87 and 88) from bird of paradise flower.
Phylogenetics analysis of P. nicotianae populations. Each branch labelled with: isolates number-host plantsgeographic origin-year of isolation-mating type. Host abbreviations—Poin: poinsettia; Lili: Lilium sp.; Als: Alstroemeria sp.; Ane: anemone; Alo: Aloe vera; Lim: limonium; Asp: asparagus; Gerb: Gerbera; Vand: Vanda sp.; Brod: Brodiaea sp.; Kln: kalanchoe; Strw: strawberry; Drac: Dracaena sp.; Lav: lavender; Boug: Bougenvillea sp.; Pine: pineapple; Tbc: tobacco; Abu: Abutilon; Echi: Echium; Prw: periwinkle; Woni: Whelsh onion; Oni: onion; Daph: daphne; Pas: passion fruit; Car: carnation; Hib: hibiscus; Rsin: Radermachera sinica; Orn: Ornithogallum; Zeb: zebra plant; Hed: Hedera (English ivy)
The isolates collected from the same host and geographic origin in different years were observed to have different genotypes. The isolates from kalanchoe in Gifu in 2004 were grouped in clades 2 and 3 (No. 10, 9, 12; and 5–8, 11, respectively), while the isolates collected in 2007 (No. 17) were found in the Clade 1. Isolates collected from Ornithogallum sp. in Chiba in 1992 (No. 80 and 82) were grouped in Clade 4, while the isolates collected in 1993 (No. 81) clustered in Clade 5. Interestingly, the isolate (No. 81) from Ornithogallum sp. had the same genotype as a parsley isolate (No. 85), although it was collected from a different host and geographic origin.
Population structure analysis
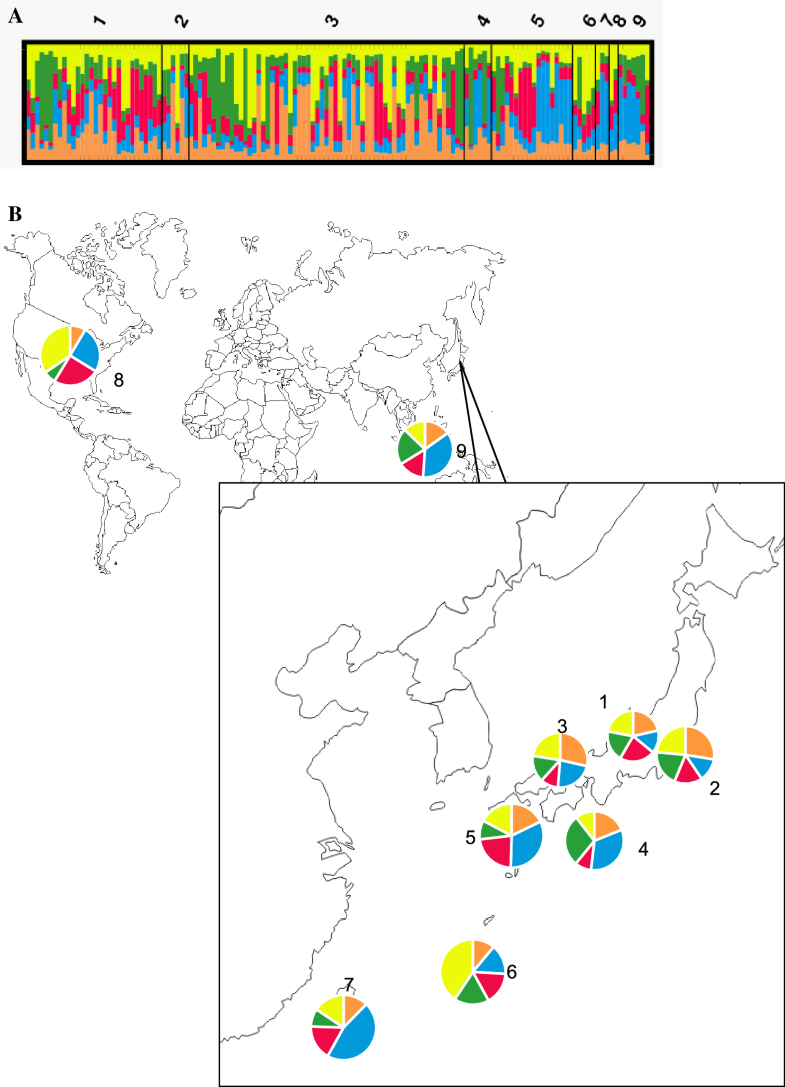
Cluster analysis revealed that the optimal number of genotypic clusters represented within the data was K = 5 and that all isolates had all clusters at different proportions. Furthermore, the populations consisted of highly admixed individuals (Fig. 2a). However, several genotypic clusters were found to be predominant in one area but minor in another. In Fig. 2, the blue cluster was prevalent in populations from Kyushu, Taiwan, and Indonesia. A fourth cluster (green) was identified in populations from Japan, Taiwan, and Indonesia but rarely in those from the United States. The yellow cluster was predominant in the southern islands and U.S. populations (Fig. 2b).
Cluster analysis of Phytophthora nicotianae using structure v. 2.3.4 (a). Genotypic clustering in each population (1: Chubu; 2: Kanto; 3: Kansai; 4: Shikoku; 5: Kyushu; 6: southern islands; 7: Taiwan; 8: USA; 9: Indonesia). b Proportions of the genotypic clusters in each population
The analysis of molecular variance (AMOVA) of microsatellite genotype data showed that isolates have low diversity among the populations (3%) but high diversity among individuals within a population (Table 4). The low Fixation Index (FST) of 0.033 with a p value of 0.082 meant that there was no significant difference between populations and a high possibility of gene flow between the populations and limited contribution of geographical origin to the genetic variance of P. nicotianae populations in Japan.
Summary of analysis of molecular variance (AMOVA) of Phytophthora nicotianae populations used in this study
| Source | df | SS | MS | Est. var. | % | Fstatistic | p value |
|---|---|---|---|---|---|---|---|
| Among populations | 8 | 28.014 | 3.502 | 0.064 | 3 | FST = 0.033 | 0.087 |
| Within populations | 267 | 500.812 | 1.876 | 1.876 | 97 | ||
| Total | 275 | 528.826 | 1.940 | 100 |
Discussion
Previous studies on the population genetics of P. nicotianae have been based on mitochondrial and nuclear DNA (Colas et al. 1998; Mammella et al. 2013). Importantly, these studies used isolates from Australia and the United States, so little was known about P. nicotianae populations from Japan. To provide a better understanding of how the pathogen is likely to emerge at a more local level in Japan, here we developed novel microsatellite markers to amplify six loci from 138 isolates from six regions in Japan and 13 isolates from overseas for comparison. A high level of polymorphism was revealed, ranging from 4 (AA-CAG) to 11 (AA-GTA) alleles per locus (Table 3). The 39 alleles amplified from six microsatellite loci is much higher than reported for P. infestans (Montarry et al. 2010) and P. sojae (Wu et al. 2017) perhaps due to the broader host range of P. nicotianae. P. capsici, which also has a wide host range, was reported to have 5–14 alleles per locus (Meitz et al. 2010), while P. alni, with a narrow host range, has as few as 2–3 per locus (Aguayo et al. 2010).
The pathogen isolated from kalanchoe in Gifu was scattered across several clades of the phylogenetic tree (Fig. 1). These isolates were collected from the same farm. In this case, the different year of isolation was a significant factor. Isolates from 2004, 2008, and 2009 were grouped into clades 2 and 3 on the phylogenetic tree, whilst the isolates from 2005 to 2007 occupied Clade 1. Novel genetic variance found in the different years of isolation could have been introduced via plant materials (such as potting mixture, seedlings, or irrigation water) because all the isolates from the previous year were type A2, thus preventing sexual recombination.
By contrast, isolates from the Saga Prefecture tended to group according to mating type and host, rather than year of isolation. Type A2 isolates from asparagus were grouped in Clade 3, even though they were collected during a different year, while type A1 was in the Clade 1. The isolates from pineapple in Indonesia were also grouped into a single clade (Clade 3) and differed from the isolates from pineapple in Taiwan (Clade 4). These results show that the sources of infection are local and specific to those host plants.
The clustering and statistical analysis revealed that P. nicotianae in Japan had high variation among individuals and a lack of geographical structure. Cluster analysis using structure showed that the P. nicotianae in Japan is highly admixed in all the isolates because there was less than 80% similarity within any one genetic cluster. This admixture could benefit the pathogen by increasing the degree of genetic variation within the population, thus raising the likelihood that novel genotypes with new combinations of traits will arise through natural selection and that deleterious mutations caused by inbreeding will be masked (Verhoeven et al. 2011). This condition is likely to be due to the choice of host plants used in this study, the majority of which were ornamental. Isolates from ornamental species are more likely to exhibit high genetic variation due to the admixtures of diverse genotypes, resulting from the trading of infected plant material between nurseries in different countries (Biasi et al. 2016).
Inconsistency between genotypic clusters and geographical origins are common in demographic analyses of Phytophthora species. Previous studies on P. nicotianae isolated on citrus (Biasi et al. 2016), P. plurivora (Schoebel et al. 2014), and P. colocasiae (Nath et al. 2013) also showed moderate to high genetic diversity without any clear relationship with the geographical origin. In the present study, the high number of genotypic clusters in a population was found to be linear to the percentage of variance among the individuals of the population. Because the AMOVA tests confirmed that variance was high within the population (97%) and low among the population (3%), while the number of genotypic clusters suggested by STRUCTURE HARVESTER was relatively high (ΔK = 5). The low number of FST value (0.033) and the associated p value of 0.08 revealed that there was no significant genetic differentiation among populations. The undifferentiated population indicates the possibility of sharing genetic materials between the populations (Ma et al. 2015), which could explain why there was no strong geographical structuring in the Japanese populations of P. nicotianae.
The lack of strong geographical structure in the P. nicotianae populations in Japan could be evidence that isolates have migrated via human activities. Since P. nicotianae is soil- and water-borne and can survive in its chlamydospore state for a long time, it could be transported via agricultural products or watercourses. Both the phylogenetic analysis and population structure results agree with a previous study in which it was hypothesized that P. nicotianae has been spread worldwide via plant material and subsequent progressive lineage diversion (Mammella et al. 2013). The pathogen was likely to respond rapidly to natural selection imposed by newly introduced host resistance genes or fungicides (Nath et al. 2013) Moreover, the ability of P. nicotianae to reproduce both sexually and asexually will enable the pathogen to be more genetically diverse. While this study has not identified the original source of P. nicotianae in Japan, it has provided a better understanding of P. nicotianae gene flow and of its evolutionary potential in Japan. Further studies should include isolates from nearby countries and improved sampling proportions to determine the route(s) of migration by P. nicotianae.
Notes
Acknowledgements
The authors acknowledge Mr. Seiji Uematsu, Dr. Hideki Watanabe, Mr. Minoru Inada, Dr. Yuji Kajitani for providing P. nicotianae isolates used in this study.
Supplementary material
References
- Aguayo J, Adams GC, Halkett F, Catal M, Husson C, Nagy ZÁ, Hansen EM, Marçais B, Frey P (2010) Strong genetic differentiation between North American and European populations of Phytophthora alni subsp. uniformis. Phytopathology 103:190–199CrossRefGoogle Scholar
- Asuyama H (1934) New diseases and pathogens reported in the year of 1934 on our cultivated plants in Japan (in Japanese). Jpn J Phytopathol 4:191–197CrossRefGoogle Scholar
- Bebber DP, Holmes T, Gurr SJ (2014) The global spread of crop pests and pathogens. Global Ecol Biogeogr 23:1398–1407CrossRefGoogle Scholar
- Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580CrossRefGoogle Scholar
- Biasi A, Martin FN, Cacciola SO, di San Lio GM, Grünwald NJ, Schena L (2016) Genetic analysis of Phytophthora nicotianae populations from different hosts using microsatellite markers. Phytopathology 106:1006–1014CrossRefGoogle Scholar
- Bonnet Ph, Lacourt I, Venard P, Ricci P (1994) Diversity in pathogenicity to tobacco and in elicitin production and isolates of Phytophthora parasitica. J Phytopathol 141:25–37CrossRefGoogle Scholar
- Bruberg MB, Elameen A, Le VH, Nærstad R, Hermansen A, Lehtinen A, Hannukkala A, Nielsen B, Hansen J, Andersson B, Yuen J (2011) Genetic analysis of Phytophthora infestans populations in the Nordic European countries reveals high genetic variability. Fungal Biol 115:335–342CrossRefGoogle Scholar
- Charlesworth B (2015) What use is population genetics? Genetics 200:667–669CrossRefGoogle Scholar
- Cline ET, Farr DF, Rossman AY (2008) A synopsis of Phytophthora with accurate scientific names, host range, and geographic distribution. Plant Health Prog. https://doi.org/10.1094/PHP-2008-0318-01-RS Google Scholar
- Colas V, Lacourt I, Ricci P, Vanlerberghe-Masutti F, Poupet A, Panabières F (1998) Diversity of virulence in Phytophthora parasitica on tobacco, as reflected by nuclear RFLPs. Phytopathology 88:205–212CrossRefGoogle Scholar
- Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361CrossRefGoogle Scholar
- Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587Google Scholar
- Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578CrossRefGoogle Scholar
- Horie H (2007) Studies on diagnosis, ecology and control of plant diseases on various horticultural crops in Japan (in Japanese). Jpn J Phytopathol 73:138–140CrossRefGoogle Scholar
- Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332CrossRefGoogle Scholar
- Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806CrossRefGoogle Scholar
- Kanto T, Uematsu S, Aino M (2007) Phytophthora blight of poinsettia (Euphorbia pulcherrima Willd. ex Klotzsch.) caused by Phytophthora nicotianae van Breda de Haan (1896) (in Japanese with English summary). Jpn J Phytopathol 73:112–113CrossRefGoogle Scholar
- Lees AK, Wattier R, Shaw DS, Sullivan L, Williams NA, Cooke DEL (2006) Novel microsatellite markers for the analysis of Phytophthora infestans populations. Plant Pathol 55:311–319CrossRefGoogle Scholar
- Ma L, Ji YJ, Zhang DX (2015) Statistical measures of genetic differentiation of populations: rationales, history and current states. Curr Zool 61:886–897CrossRefGoogle Scholar
- Mammella MA, Martin FN, Cacciola SO, Coffey MD, Faedda R, Schena L (2013) Analyses of the population structure in a global collection of Phytophthora nicotianae isolates inferred from mitochondrial and nuclear DNA sequences. Phytopathology 103:610–622CrossRefGoogle Scholar
- Matsuzaki M (1988) Distribution of mating types of Phytophthora nicotianae var. parasitica, causal fungus of Phytophthora rot of strawberry, in Saga Prefecture (in Japanese with English summary). Ann Phytopathol Soc Jpn 54:544–547CrossRefGoogle Scholar
- Meitz J, Linde C, Thompson A, Langenhoven SD, McLeod A (2010) Phytophthora capsici on vegetable hosts in South Africa: distribution, host range and genetic diversity. Australas Plant Pathol 39:431–439CrossRefGoogle Scholar
- Montarry J, Andrividon D, Glais I, Corbiere R, Mialdea G, Delmotte F (2010) Microsatellite markers reveal two admixed genetic groups and an ongoing displacement within the French population of the invasive plant pathogen Phytophthora infestans. Mol Ecol 19:1965–1977CrossRefGoogle Scholar
- Morita Y, Tojo M (2007) Modification of PARP medium using fluazinam, miconazole, and nystatin for detection of Pythium spp. in soil. Plant Dis 91:1591–1599CrossRefGoogle Scholar
- Nakamura H, Matsuzaki M (1994) Occurrence of Phytophthora rot of limonium caused by Phytophthora nicotianae in Saga Prefecture (Abstract in Japanese). Ann Phytopathol Soc Jpn 60:737Google Scholar
- Nath VS, Senthil M, Hegde VM, Jeeva ML, Misra RS, Veena SS, Raj M (2013) Genetic diversity of Phytophthora colocasiae isolates in India based on AFLP analysis. 3 Biotech 3:297–305CrossRefGoogle Scholar
- Panabières F, Ali GS, Allagui MB, Dalio RJD, Gudmestad NC, Kuhn ML, Guha Roy S, Schena L, Zampounis A (2016) Phytophthora nicotianae diseases worldwide: new knowledge of a long-recognised pathogen. Phytopathol Mediterr 55:20–40Google Scholar
- Parkunan V, Johnson CS, Bowman BC (2010) Population structure, mating type, and mefenoxam sensitivity of Phytophthora nicotianae in Virginia tobacco fields. Plant Dis 94:1361–1365CrossRefGoogle Scholar
- Peakall R, Smouse PE (2006) GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Resour 6:288–295CrossRefGoogle Scholar
- Peakall R, Smouse PE (2012) GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539CrossRefGoogle Scholar
- Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 7:574–578Google Scholar
- Robideau GP, De Cock AW, Coffey MD, Voglmayr H, Brouwer H, Bala K, Chitty DW, Désaulniers N, Eggertson QA, Gachon CM, Hu CH, Küpper FC, Rintoul TL, Sarhan E, Verstappen EC, Zhang Y, Bonants PJ, Ristaino JB, Lévesque CA (2011) DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol Ecol Resour 11:1002–1011CrossRefGoogle Scholar
- Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138CrossRefGoogle Scholar
- San Millán RM, Martínez-Ballesteros I, Rementeria A, Garaizar J, Bikandi J (2013) Online exercise for the design and simulation of PCR and PCR-RFLP experiments. BMC Res Notes 6:513. https://doi.org/10.1186/1756-0500-6-513 CrossRefGoogle Scholar
- Schoebel CN, Stewart J, Gruenwald NJ, Rigling D, Prospero S (2014) Population history and pathways of spread of the plant pathogen Phytophthora plurivora. PLOS One 9:e85368CrossRefGoogle Scholar
- Selkoe KA, Toonen RJ (2006) Microsatellites for ecologist: a practical guide to using and evaluating microsatellite markers. Ecol Lett 9:615–629CrossRefGoogle Scholar
- Sunnucks P (2000) Efficient genetic markers for population biology. Trends Ecol Evol 15:199–203CrossRefGoogle Scholar
- Suzui T, Makino T, Ogoshi A (1980) Phytophthora rot of strawberry caused by Phytophthora nicotianae var. parasitica in Shizuoka. Ann Phytopathol Soc Jpn 46:169–178CrossRefGoogle Scholar
- Takeuchi J, Horie H (2000) First report of Phytophthora rot of garden pea and Albuca nelsonii in Japan (in Japanese with English summary). Annu Rept Kanto-Tosan Plant Prot Soc 47:45–48Google Scholar
- Takeuchi T, Suzuki T (2010) Phytophthora blight (Phytophthora nicotianae) on hydroponically grown Welsh onion (Allium fistulosum L.) and controlling damage with the nutrient solution (in Japanese with English summary). Bull Chiba Agric Res Cent 2:1–6Google Scholar
- Takeuchi J, Horie H, Eimori K (2004) First report of Phytophthora rot of New Zealand spinach in Japan (in Japanese with English summary). Annu Rept Kanto-Tosan Plant Prot Soc 51:55–57Google Scholar
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729CrossRefGoogle Scholar
- Tashiro N, Uematsu S, Matsuzaki M, Ide Y, Etoh T (2002) Phytophthora palmivora, P. citrophthora and P. nicotianae as causal agents of citrus brown rot (Abstract in Japanese). Jpn J Phytopathol 68:189Google Scholar
- Verhoeven KJF, Macel M, Wolfe LM, Biere A (2011) Population admixture, biological invasions and the balance between local adaptation and inbreeding depression. Proc R Soc B 278:2–8CrossRefGoogle Scholar
- Watanabe H, Taguchi Y, Hyakumachi M, Kageyama K (2007) Pythium and Phytophthora species associated with root and stem rots of kalanchoe. J Gen Plant Pathol 73:81–88CrossRefGoogle Scholar
- Wu M, Li B, Liu P, Weng Q, Zhan J, Chen Q (2017) Genetic analysis of Phytophthora sojae populations in Fujian, China. Plant Pathol 66:1182–1190CrossRefGoogle Scholar
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform 13:134CrossRefGoogle Scholar
- Yokota S, Oomori T, Nao M, Watanabe T, Kitamoto H (2013) Involvement of Phytophthora rot caused by Phytophthora nicotianae in growth failure of asparagus (Asparagus officinalis L.) in replanted fields in Ehime Prefecture. Soil Microorg 67:77–82Google Scholar